Background and Overview
In the past few years, deep learning foundation models have been trendy, especially in the computer vision and natural language processing. As a result, many milestone works have been proposed, such as Vision Transformers (ViT), Generative Pretrained Transformer (GPT), and Contrastive Language-Image Pretraining (CLIP) . They aim to solve many downstream tasks by utilizing the robust representation learning and generalization abilities of foundation models.
However, the lack of public availability and quality annotations in medical imaging has been the bottleneck for training large-scale deep learning models for many downstream clinical applications. It remains a tedious and time-consuming job for medical professionals to hand-label volumetric data repeatedly while providing a few differentiable sample cases is more logically feasible and complies with the training process of medical residents. In this case, a foundation model, often trained with thousands of millions of images and probably other modalities of data, could serve as the base for building applications with a single or a few cases in the form of prompt learning.
Thus, we propose holding the challenge of foundation models for medical image analysis in conjunction with Grand Challenge. This challenge aims at revealing the power of foundation models to ease the effort of obtaining quality annotations and target improving the classification accuracy of tail classes. It aligns with the recent trend and success of building foundation models mentioned above for a variety of downstream applications. It is designed to promote the foundation model and its application in solving frontline clinical problems in a setting where only one or a few sample cases (with quality annotations) are available during downstream task adaptation. It also fits perfectly for the long-tailed classification scenario, with only a few rare disease cases available for the training. In a word, our challenge aims to advance technique in prompting large-scale pre-trained foundation models via a few data samples as a new paradigm for medical image analysis, e.g., classification tasks proposed here as use cases.
In the challenge, during the training phase, a small amount of our private data would be utilized for the initial training (selected few samples) and validation (the rest of the dataset). Participants are encouraged to achieve higher performance scores in three detailed application tasks, i.e., thoracic disease screening in chest radiography, pathological tumor tissue classification, and lesion classification in colonoscopy images. The final evaluation will be conducted in the same setting on the reserved private datasets, i.e., the random selection of a few samples for the training and the rest for the testing. The final metrics will be averaged over five individual runs of the same prompting/testing process.
We would provide code/docker examples that include the pipeline of this challenge, such as model baseline (using popular pre-trained foundation models in computer vision), dataset preparation, and model evaluation. The participants will need to focus on improving the overall performance of their model adaptation approaches on all three tasks.
To complement the challenge, we also organize a special issue in the prestigious journal, Medical Image Analysis, supported by Chief Editors, with Prof. Dimitris Metaxas as the lead editor. The winners of the challenge will be invited to submit their groundbreaking solutions in a summarization paper, which will be a valuable resource for the entire medical imaging analysis community.
Tasks
Here are three main tasks.
Task 1:
Thoracic Disease Screening
Chest X-ray is a regularly adopted imaging technique for daily clinical routine. Many thoracic diseases are reported, and further examinations are recommended for differential diagnoses. Due to the large amount and fast reporting requirements in certain emergency facilities, swift screening and reporting of common thoracic diseases could largely improve the efficiency of the medical process. Although there are a few chest x-ray datasets (e.g., ChestX-ray14, Chexpert, MIMIC-CXR) now publicly accessible, images with quality annotations (preferably verified by radiologists) are still a scarce resource for training and evaluating modern machine learning-based models.
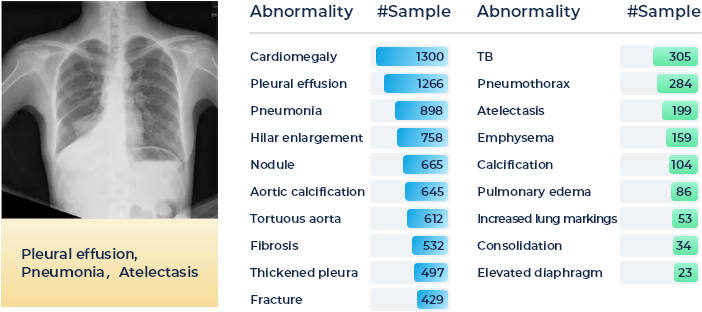
In this task, the participants shall target providing an accurate thoracic disease screening of 19 common abnormalities by utilizing the provided foundation model and following the one-shot/few-shot learning setting, e.g., with 1, 5, or 10 sample cases for each disease category. A detailed distribution of 19 common thoracic diseases is presented in the Figure below, which is sorted by the number of samples. Tail classes are highlighted in Green. A total of 2,140 patients' 2,140 frontal radiography images will be provided as the training set and a total of 2,708 patients' 2,708 frontal radiography images will be provided as the validation set. Each PNG image is converted from the original DICOM files using the default window level and width (stored in the DICOM tags).
Task 2:
Pathological Tumor Tissue Classification
In clinical diagnosis, quantifying cancer cells and regions is the primary goal for pathologists. Pathology examination can find cells of early-stage tumors from small tissue slices. Approaches for the classification of pathological tissue patches are desired to ease this process and help screen whether regions of malignant cells are in the entire slide in a sliding window manner. In pathologists' daily routine, they need to examine hundreds of tissue slices, which is a time-consuming and exhausting job.
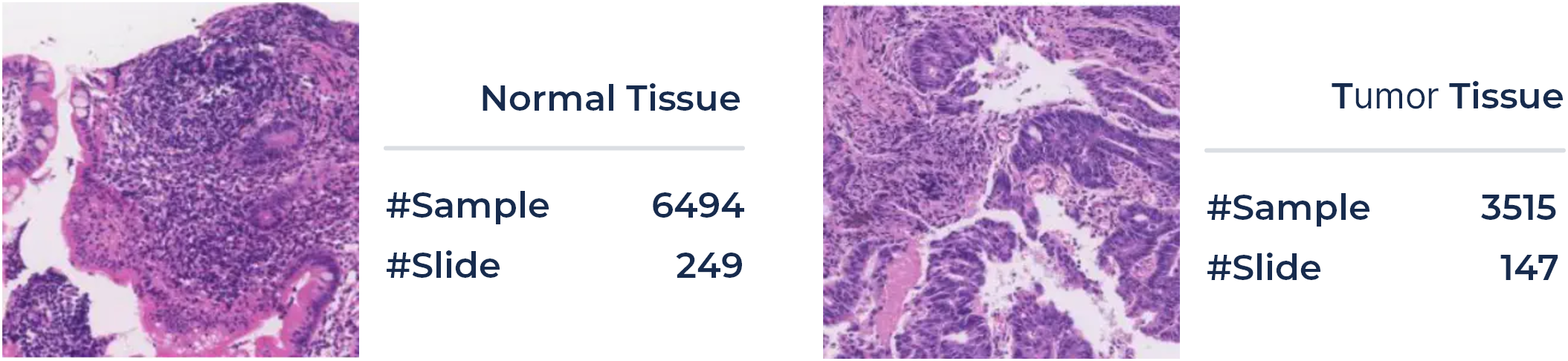
Here we propose a challenging task on the automatic classification of pathological tissue patches, aiming at an automatic classification of the large patches. For the training and validation phase, a total of 396 patients' 10,009 large tissue patches (4,654 and 5,355 patches for training and validation phase, with a uniform size of 1024x1024, respectively) of colonoscopy pathology examination will be available in ColonPath.Positive and negative patch samples (with and without tumor tissue) are illustrated in the Figure below, along with the number of samples in each category. Tissue patches are extracted from the WSI in a sliding window fashion with a fixed size of 1024x1024 and a stride of 768. Following the one-shot/few-shot learning setting, e.g., with 1, 5, or 10 patients for each category, a classification model of tumor tissue patches will be developed based on the provided foundation model. The rest of the data will be employed for the validation of the tuned model. In a similar setting, another 469 patients' 10,650 large tissue patches will be utilized in the evaluation phase.
Task 3:
Lesion Classification in Colonoscopy Images
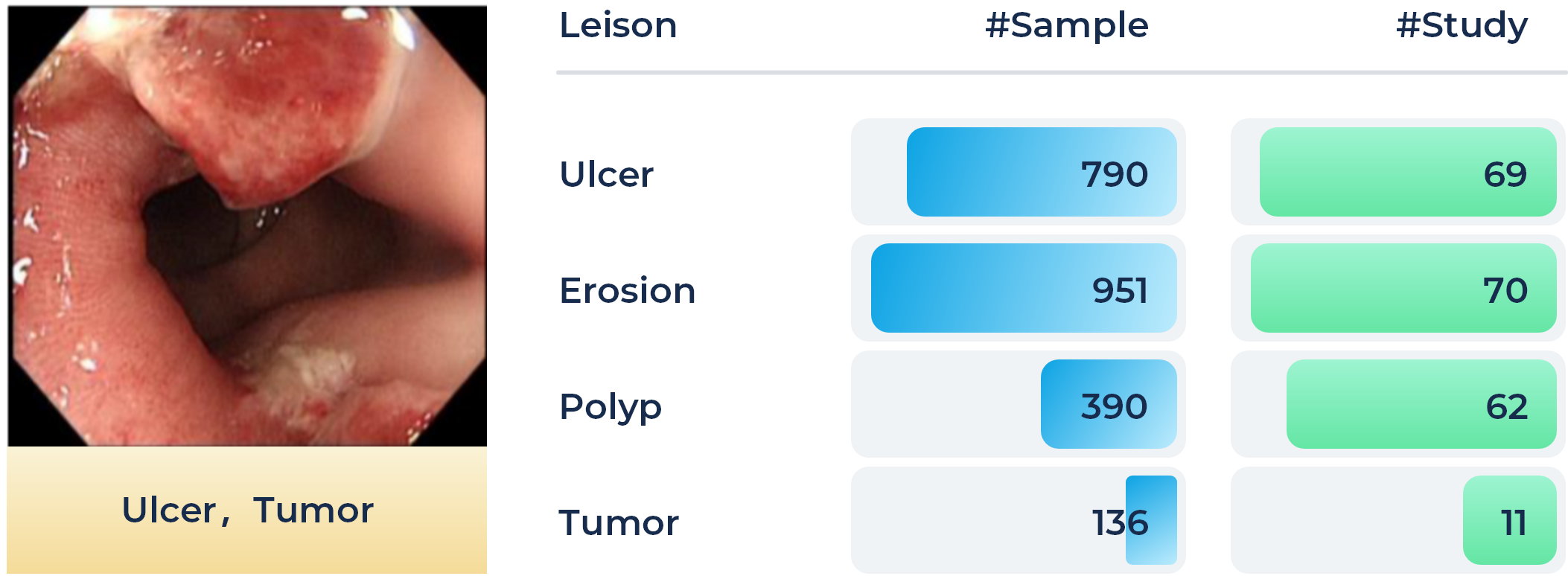
Colorectal cancer is one of the most common and fatal cancers among men and women around the world. Abnormalities like polyps and ulcers are precursors to colorectal cancer and are often found in colonoscopy screening of people aged above 50. The risks largely increase along with aging. Colonoscopy is the gold standard for the detection and early diagnosis of such abnormalities with necessary biopsy on site, which could significantly affect the survival rate of colorectal cancer. Automatic detection of such lesions during the colonoscopy procedure could prevent missing lesions and ease the workload of gastroenterologists in colonoscopy.
In this task, the participants shall target providing an accurate classification of four different lesion types by utilizing the provided foundation model and following the one-shot/few-shot learning setting, e.g., with 1, 5, or 10 sample cases for each lesion category. A total of 80 patients' 3865 images (1,810 and 2,055 images for training and validation phase, with an average size of 1280 x 1024, respectively.) recorded during the colonoscopy examination on the workstations are collected as the training set. Four types of lesions, i.e., ulcer, erosion, polyp, and tumor, are included, which are illustrated in the Figure below, along with the number of samples in each category.Additionally, another 2,199 colonoscopic images will be collected from another hospital for the final evaluation.
Organizers
Technical Committee
Dequan Wang
Shanghai Jiao Tong University, Shanghai, China.
Xiaosong Wang
Shanghai AI Laboratory, Shanghai, China.
Qi Dou
The Chinese University of Hong Kong, Hongkong, China
Xiaoxiao Li
The University of British Columbia, Vancouver, Canada.
Dimitris Metaxas
Rutgers University, New Jersey, USA.
Shaoting Zhang
Shanghai AI Laboratory, Shanghai, China.
Clinical Committee
Qian Da
Shanghai Ruijin Hospital, Shanghai, China.
Fangfang Cui
The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, China.
Feng Gao
The Sixth Affiliated Hospital of Sun Yat-sen University, Guangzhou, China
Jun Shen
Renji Hospital, Shanghai, China.
Kang Li
West China Hospital, Sichuan University, Chengdu, China.
Webmaster
Mengzhang Li
Shanghai AI Laboratory, Shanghai, China.
FAQ
Timeline of this Challenge
Pleaser refer to https://medfm2023.grand-challenge.org/ for more details.
Submission of this Challenge
In the validation phase, participants will expect to submit prediction results on validation set directly to the submission site.
In the tesing phase, participants will expect to submit docker container (code and foundation model) to the submission site.
Pleaser refer to https://medfm2023.grand-challenge.org/ for more details.
Who should I contact if I have any questions?
You should contact the liaison by emailing openmedlab@pjlab.org.cn.
Related Workshops
MedAGI
MICCAI 2023 1st International Workshop on Foundation Models for General Medical AI.
https://medagi.github.io/#/
Sponsors
Would like to join the community and improve your tech brand?
Contact Us








